Traditional medicinal chemistry has focused on single-target drugs, effective for many conditions but inadequate for complex diseases like cancer. Polytherapy tackles this but faces challenges. Multi-Target Directed Ligands (MTDLs) offer a novel approach, targeting multiple disease pathways for enhanced efficacy. Integrating MTDLs with lipidomics shows promise in GB therapy development.
Key words: lipid biology, multi target treatment, brain tumors, personalised medicine
Introduction
The traditional pharmaceutical approach of designing a disease/symptom-specific drug has historically proven effective. However, this methodology often falls short due to the complex and often multifactorial etiology of diseases, such as cancer.[1, 2] Moreover, prolonged use of certain antineoplastic drugs can lead to drug resistance and reduced efficacy over time, ultimately diminishing the overall therapeutic effect.[3, 4] Polytherapy, defined as the simultaneous administration of multiple drugs in the form of a drug cocktail to target various disease symptoms, is often the approach to treat multifactorial diseases such as cancer.[5, 6] Considering that single-target therapeutics are developed independently of each other, implementation of polytherapy often presents challenges due to unpredictable drug-drug interactions due to unknown pharmacological profiles. The resulting drug-drug interaction can lead to unwanted adverse effects and disqualify the drug cocktail from therapeutic use.
Multi-Target Directed Ligands and Drug Discovery
Due to these limitations, there’s been a shift from the traditional single-target drug strategy towards a new approach known as Multi-Target Directed Ligands (MTDLs), also referred to as polypharmacology (Fig. 1). Unlike polytherapy, which uses multiple therapeutics to treat different targets of the same disease independently, polypharmacology designs single therapeutics that act on multiple targets or disease pathways.[7] The MTDL strategy allows for the attenuation of multiple interconnected pathways, potentially resulting in greater therapeutic efficacy than the sum of individual single-target drugs.[8] MTDLs often exhibit synergistic effects, i.e. enhancing efficacy by targeting multiple sites simultaneously, allowing for the design of less potent MTDLs that produce comparable outcomes.[9] Apart from the traditional challenges faced by single-target drugs, the nature of the disease pathology has a significant impact on the therapeutic efficacy of a drug.
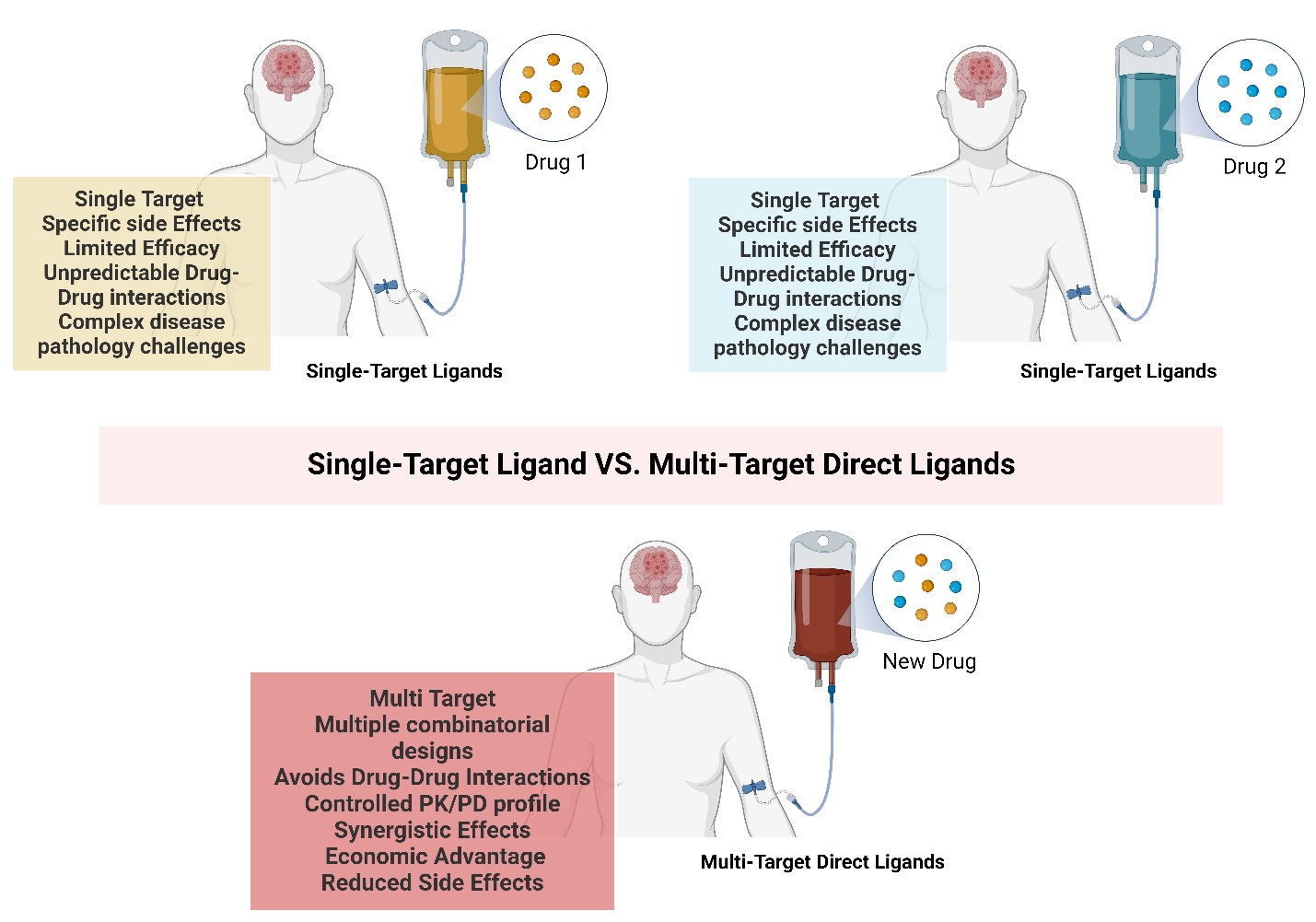
Figure 1. Comparison of single drug strategy and MTDL strategy. Single-target drugs are represented in pink and blue. MTDL drug is represented in purple.
Furthermore, the complex pathology of such diseases limits a drug's ability to mitigate disease progression. The single-target strategy alleviates a single aspect or rather symptoms of the disease, however, the alternative or compensatory cellular processes will continue to contribute to the overall progression of the disease. This is particularly evident in cancer treatment, where the initial effects of therapy may be negated by the activation of alternative survival pathways in cancerous cells, ultimately leading to relapse.
To summarize, MTDLs offer several advantages over single-target approaches:
1. Increased efficacy: By targeting multiple signaling pathways, MTDLs can have a broader therapeutic impact and reduce the likelihood of disease evading treatment through compensatory mechanisms.
2. Lower resistance: Targeting multiple pathways simultaneously makes it more difficult to develop resistance in diseases such as cancer, as multiple mutations or adaptations would be required.
3. Synergistic effects: MTDLs can achieve greater therapeutic effects than the sum of their parts, as the simultaneous modulation of multiple targets can lead to synergistic interactions that increase efficacy.
4. Network pharmacology: This approach leverages the growing understanding of complex cellular networks, enabling the development of drugs that can modulate multiple nodes within these networks for a broader therapeutic effect.
The development of MTDL involves sophisticated drug development techniques. Techniques such as machine learning are employed to predict the effects of interfering with various physiological pathways, which can then be further integrated with data collected from a variety of ‘omics’ technology disciplines such as genomics, proteomics, and lipidomics [10]. This holistic approach allows for more precise and targeted development of therapeutics, providing hope for more effective treatments for complex diseases that are difficult to treat with traditional single-target drugs.
MTDLs can be effectively utilised in conjunction with multi-omics approaches for drug discovery by integrating genomic, transcriptomic, proteomic, lipidomic, and metabolomic data to identify multiple therapeutic targets involved in complex diseases [11]. Multi-omics provides a comprehensive understanding of disease mechanisms, revealing interconnected pathways and potential target networks [12]. MTDLs are then designed to modulate these targets simultaneously, enhancing therapeutic efficacy and reducing side effects [13, 14]. This synergy accelerates the identification of promising drug candidates, optimises their design, and allows for a more precise and personalised treatment strategy, ultimately improving drug development outcomes [15-17]. In the following section, we summarize the cross-application of Lipidomics and MTDL for new drug discovery in glioblastoma (GB).
MTDL and GB New Drug Discovery Using Lipidomics Approach
Current anticancer therapies for GB include TMZ and other alkylating agents, often involving polytherapy to increase efficacy and avoid resistance.[18, 19] The MTDL strategy could represent a new avenue for developing novel anti-GB therapeutics that target two or more GB specific targets, potentially overriding the resistance that is commonly associated with TMZ therapy. The first step is to recognise novel targets, and data obtained from e.g. lipidomics may represent an excellent source for target identification (Fig. 2). Lipidomics, the large-scale study of cellular lipid pathways and networks using advanced analytical techniques like mass spectrometry, provides an excellent source for identifying novel targets.
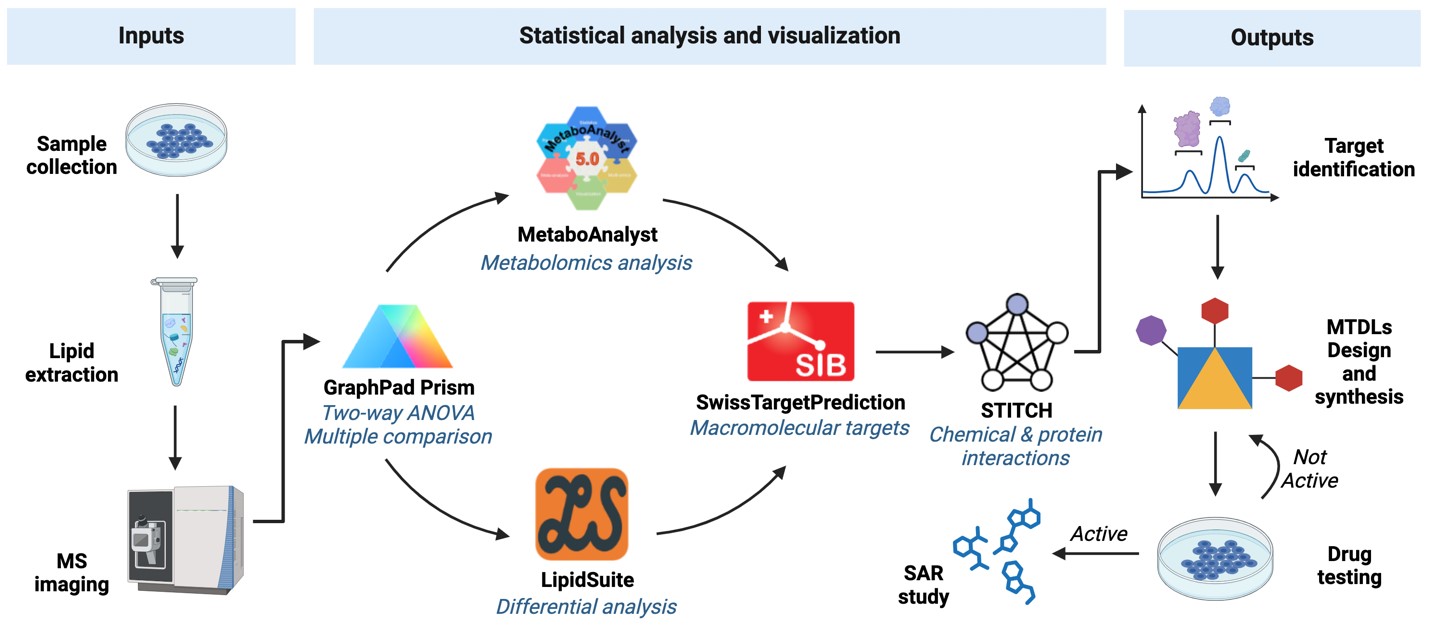
Figure 2. Example workflow with MS-based data and selected lipidomics tools aiding the discovery of novel targets for MTDLs design and synthesis.
Traditional drug discovery processes rely on high-throughput screening (HTS) commonly complemented with virtual-ligand screening (VLS) and X-ray crystallography. Despite HTS’ strength to identify a novel lead compound with therapeutic potential, HTS cannot be used to identify the novel target(s). In addition to the familiar functions of lipids such as energy storage, cellular membrane, and signal transduction, lipids can also be acquired by cancer cells to undergo metabolic reprogramming and contribute to the survival of cancerous cells.[20] This metabolic reprogramming results in the alternative expression of lipid metabolising enzymes, which then serves as the hallmark of various pathologies.[21] For example, changes in lipid levels have been reported in obesity, atherosclerosis, type II diabetes, and metabolic syndromes.[20, 22] Lipidomics represents the large-scale analysis of lipids using mass-spectrometry as the analytical method to identify and quantify cellular lipid levels.[23] In addition, this approach utilises the extensive profiling capabilities of lipidomics to uncover unique metabolic alterations specific to GB. By identifying lipid-related biomarkers and metabolic pathways that are altered in GB, we can better understand the pathophysiology of the disease and develop more targeted therapeutic strategies. Integrating lipidomic data into MTDL design not only improves target identification, but also the precision and efficacy of therapeutics.
The proposed workflow begins with the collection and preparation of biological samples from GB tissue, followed by lipid extraction and separation using advanced mass spectrometry techniques. Analysing the resulting data with specialised bioinformatics tools will then identify differentially expressed lipid species and their associated metabolic pathways. This information is crucial for identifying new target molecules and understanding their role in the progression of GB.
Once new targets are identified, the development of MTDLs will combine pharmacophoric elements that can interact with multiple targets simultaneously, aiming to disrupt multiple critical GB metabolic pathways. This multi-targeted approach may bear the potential to overcome the limitations of single-targe therapies and reduce the likelihood of drug resistance.
A key component of this workflow is the integration of structural analyses, including techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM). These techniques provide high-resolution data on the structure of target proteins and their complexes with potential MTDLs.
The structural analysis facilitates the following steps:
1. Identification of the binding site: Detailed structural information helps to identify precise binding sites on the target proteins and ensures that the designed pharmacophore elements fit precisely and interact optimally with the target proteins.
2. Molecular docking and simulation: Computational methods such as molecular docking and molecular dynamics simulations are used to model the interactions between the MTDLs and their target proteins. These simulations allow the prediction of the binding affinities and stability of the drug-target complexes and serve as a basis for the optimisation of the MTDLs.
3. Structure-activity Relationship Studies (SAR): The knowledge gained from structural analysis is incorporated into SAR studies in which modifications to pharmacophore elements are tested to improve their efficacy, specificity, and safety. Structural data helps to rationalise the observed effects of different chemical modifications and to develop more effective and selective MTDLs.
Testing of the synthesized MTDLs in GB cell lines and animal models will provide initial efficacy and safety data. Successful candidates will undergo further SAR studies to optimise their pharmacological profile and ensure maximum therapeutic benefit with minimal adverse effects. The flexibility of the MTDL design allows for iterative refinement based on experimental feedback, ultimately leading to tailored and more effective therapies for GB.
In addition, combining existing drugs such as TMZ or OTX015 with newly identified targets can lead to synergistic effects that increase overall treatment efficacy. This dual approach not only expands the therapeutic arsenal against GB, but also provides a framework for the development of personalised medicine strategies tailored to the unique lipidomic profiles of individual patients.
The integration of lipidomics with MTDL design and structural analysis holds promise for advancing GB therapy. By targeting multiple pathways simultaneously and using comprehensive lipidomic and structural data, we can develop more effective Consistent, and personalised treatments for this aggressive cancer. This holistic approach represents a significant advancement in the field of drug development and offers new hope for patients with GB and other complex diseases.
Future Direction
The future of pharmaceutical development is poised for transformative advancements through the integration of MTDLs and multi-omics approaches in drug discovery. By harnessing the power of comprehensive datasets from genomics, transcriptomics, proteomics, and metabolomics, researchers can gain a holistic understanding of disease pathways and their complex interactions. This multi-omics perspective enables the identification of multiple key therapeutic targets involved in multifactorial diseases such as cancer, neurodegenerative disorders, and metabolic syndromes.
The design of MTDLs to simultaneously modulate these targets can lead to more effective and synergistic treatments, addressing the root causes of the disease rather than just alleviating symptoms. This approach also holds promise in overcoming drug resistance, a significant challenge in prevailing therapies.
Future directions will focus on machine learning and artificial intelligence to integrate and analyse multi-omics data, predict optimal target combinations, and design novel MTDLs. Additionally, personalised medicine will benefit from this integration, as patient-specific omics data can guide the development of tailored MTDLs, maximising therapeutic efficacy while minimising adverse effects. This convergence of technologies promises a new era of precision and efficacy in drug discovery and development.
References
1. Reddy, A.S. and S. Zhang, Polypharmacology: drug discovery for the future. Expert Rev Clin Pharmacol, 2013. 6(1): p. 41-7.
2. Lillich, F.F., J.D. Imig, and E. Proschak, Multi-Target Approaches in Metabolic Syndrome. Front Pharmacol, 2020. 11: p. 554961.
3. Housman, G., et al., Drug resistance in cancer: an overview. Cancers (Basel), 2014. 6(3): p. 1769-92.
4. Mansoori, B., et al., The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv Pharm Bull, 2017. 7(3): p. 339-348.
5. Morphy, R. and Z. Rankovic, Designed Multiple Ligands. An Emerging Drug Discovery Paradigm. Journal of Medicinal Chemistry, 2005. 48(21): p. 6523-6543.
6. Ryszkiewicz, P., B. Malinowska, and E. Schlicker, Polypharmacology: promises and new drugs in 2022. Pharmacol Rep, 2023. 75(4): p. 755-770.
7. Kabir, A. and A. Muth, Polypharmacology: The science of multi-targeting molecules. Pharmacological Research, 2022. 176: p. 106055.
8. Calzetta, L. and C. Koziol-White, Pharmacological interactions: Synergism, or not synergism, that is the question. Curr Res Pharmacol Drug Discov, 2021. 2: p. 100046.
9. Proschak, E., H. Stark, and D. Merk, Polypharmacology by Design: A Medicinal Chemist's Perspective on Multitargeting Compounds. J Med Chem, 2019. 62(2): p. 420-444.
10. Hemmati, S. and H. Rasekhi Kazerooni, Polypharmacological Cell-Penetrating Peptides from Venomous Marine Animals Based on Immunomodulating, Antimicrobial, and Anticancer Properties. Mar Drugs, 2022. 20(12).
11. Hajjo, R., et al., A Review of the Recent Advances in Alzheimer's Disease Research and the Utilization of Network Biology Approaches for Prioritizing Diagnostics and Therapeutics. Diagnostics (Basel), 2022. 12(12).
12. Ma, Y., et al., Spatial Multi-Omics in Alzheimer's Disease: A Multi-Dimensional Approach to Understanding Pathology and Progression. Curr Issues Mol Biol, 2024. 46(5): p. 4968-4990.
13. Qiu, Y. and F. Cheng, Artificial intelligence for drug discovery and development in Alzheimer's disease. Curr Opin Struct Biol, 2024. 85: p. 102776.
14. da Silva Rosa, S.C., et al., A Lipidomics Approach to Determine the Role of Lipids and Its Crosstalk with Autophagy in Lung Cancer Metastasis. Methods Mol Biol, 2024.
15. Jose, A., et al., Integration of pan-omics technologies and three-dimensional in vitro tumor models: an approach toward drug discovery and precision medicine. Mol Cancer, 2024. 23(1): p. 50.
16. Nam, Y., et al., Harnessing Artificial Intelligence in Multimodal Omics Data Integration: Paving the Path for the Next Frontier in Precision Medicine. Annu Rev Biomed Data Sci, 2024.
17. da Silva Rosa, S.C., et al., Prioritization of genes for translation: a computational approach. Expert Rev Proteomics, 2024. 21(4): p. 125-147.
18. Jacobs, J., et al., The role of BCL2L13 in glioblastoma: turning a need into a target. Biochem Cell Biol, 2024. 102(2): p. 127-134.
19. Barzegar Behrooz, A., et al., Integrating Multi-Omics Analysis for Enhanced Diagnosis and Treatment of Glioblastoma: A Comprehensive Data-Driven Approach. Cancers (Basel), 2023. 15(12).
20. Butler, L.M., et al., Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Advanced Drug Delivery Reviews, 2020. 159: p. 245-293.
21. Jin, H.-R., et al., Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics. Journal of Hematology & Oncology, 2023. 16(1).
22. Wang, M., et al., Novel advances in shotgun lipidomics for biology and medicine. Progress in Lipid Research, 2016. 61: p. 83-108.
23. Züllig, T., M. Trötzmüller, and H.C. Köfeler, Lipidomics from sample preparation to data analysis: a primer. Analytical and Bioanalytical Chemistry, 2020. 412(10): p. 2191-2209.